Figshare API Resources
You can use the Figshare API to automate record management, gather statistics, or even build custom web apps for users or administrators.
Note: Interested in migrating records in batch? This is certainly possible through the API. However, if you are a repository manager at an institution, the Batch Management Tool may be an easier option for you.
On this page:
- Advice and guidance
- Basic examples
- Retrieve information for multiple items
- Example workflows
- Create an author report
- Create records, upload files, and publish
- Batch format and upload metadata from a source
- Delete all or some of the items in an account (useful when testing in stage or if you have lots of uneeded draft records.
- Administrator workflows and reporting
- Custom Web Applicaitons
Advice and Guidance
- Understanding the API documentation
- Formatting JSON inputs and outputs
- A variety of other scripts can be found here and this help page links to Google Colab versions of some of those scripts
- There are many other GitHub repos for Figshare related workflows or integrations
- More on stats API endpoints
Examples
Retrieve data
Any public metadata or files can be retrieved through the API without authentication. In the example below, the full metadata record for an item is retrieved. The ITEM_ID is the number at the end of any item’s URL.
Python:
import json
import requests
#Set the base URL and ITEM_ID
BASE_URL = 'https://api.figshare.com/v2'
ITEM_ID = 123456
#Retrieve public metadata from the endpoint
s=requests.get(BASE_URL + '/articles/' + str(ITEM_ID))
#Load the metadata as JSON
metadata=json.loads(s.text)
#View the metadata
print(json.dumps(metadata, indent=2))
Output:
{
"files": [
{
"id": 3074543,
"name": "2011-02-15_14-24-56-228.png",
"size": 470683,
"is_link_only": false,
"download_url": "https://ndownloader.figshare.com/files/3074543",
"supplied_md5": "eb5c9c6b278b533f98aa02feae57c266",
"computed_md5": "eb5c9c6b278b533f98aa02feae57c266"
}
],
"custom_fields": [],
"authors": [
{
"id": 155524,
"full_name": "Yunyan Deng",
"is_active": false,
"url_name": "_",
"orcid_id": ""
},
{
"id": 155526,
"full_name": "Jianting Yao",
"is_active": false,
"url_name": "_",
"orcid_id": ""
},
{
"id": 155529,
"full_name": "Xiuliang Wang",
"is_active": false,
"url_name": "_",
"orcid_id": ""
},
{
"id": 110581,
"full_name": "Hui Guo",
"is_active": false,
"url_name": "_",
"orcid_id": ""
},
{
"id": 155532,
"full_name": "Delin Duan",
"is_active": false,
"url_name": "_",
"orcid_id": ""
}
],
"figshare_url": "https://plos.figshare.com/articles/dataset/Transcriptome_Sequencing_and_Comparative_Analysis_of_Saccharina_japonica_Laminariales_Phaeophyceae_under_Blue_Light_Induction/123456",
"description": "<div><h3>Background</h3><p>Light has significant effect on the growth and development of <em>Saccharina japonica</em>, but there are limited reports on blue light mediated physiological responses and molecular mechanism. In this study, high-throughput paired-end RNA-sequencing (RNA-Seq) technology was applied to transcriptomes of <em>S. japonica</em> exposed to blue light and darkness, respectively. Comparative analysis of gene expression was designed to correlate the effect of blue light and physiological mechanisms on the molecular level.</p> <h3>Principal Findings</h3><p>RNA-seq analysis yielded 70,497 non-redundant unigenes with an average length of 538 bp. 28,358 (40.2%) functional transcripts encoding regions were identified. Annotation through Swissprot, Nr, GO, KEGG, and COG databases showed 25,924 unigenes compared well (E-value <10<sup>\u22125</sup>) with known gene sequences, and 43 unigenes were putative BL photoreceptor. 10,440 unigenes were classified into Gene Ontology, and 8,476 unigenes were involved in 114 known pathways. Based on RPKM values, 11,660 (16.5%) differentially expressed unigenes were detected between blue light and dark exposed treatments, including 7,808 upregulated and 3,852 downregulated unigenes, suggesting <em>S. japonica</em> had undergone extensive transcriptome re-orchestration during BL exposure. The BL-specific responsive genes were indentified to function in processes of circadian rhythm, flavonoid biosynthesis, photoreactivation and photomorphogenesis.</p> <h3>Significance</h3><p>Transcriptome profiling of <em>S. japonica</em> provides clues to potential genes identification and future functional genomics study. The global survey of expression changes under blue light will enhance our understanding of molecular mechanisms underlying blue light induced responses in lower plants as well as facilitate future blue light photoreceptor identification and specific responsive pathways analysis.</p> </div>",
"funding": null,
"funding_list": [],
"version": 1,
"status": "public",
"size": 470683,
"created_date": "2012-06-27T00:57:36Z",
"modified_date": "2016-01-19T09:09:40Z",
"is_public": true,
"is_confidential": false,
"is_metadata_record": false,
"confidential_reason": "",
"metadata_reason": "",
"license": {
"value": 1,
"name": "CC BY 4.0",
"url": "https://creativecommons.org/licenses/by/4.0/"
},
"tags": [
"transcriptome",
"sequencing",
"comparative",
"induction"
],
"categories": [
{
"id": 69,
"title": "Inorganic Chemistry",
"parent_id": 38,
"path": "",
"source_id": "",
"taxonomy_id": 10
},
{
"id": 13,
"title": "Genetics",
"parent_id": 48,
"path": "",
"source_id": "",
"taxonomy_id": 10
},
{
"id": 61,
"title": "Developmental Biology",
"parent_id": 48,
"path": "",
"source_id": "",
"taxonomy_id": 10
}
],
"references": [],
"has_linked_file": false,
"citation": "Deng, Yunyan; Yao, Jianting; Wang, Xiuliang; Guo, Hui; Duan, Delin (2016): Transcriptome Sequencing and Comparative Analysis of <em>Saccharina japonica</em> (Laminariales, Phaeophyceae) under Blue Light Induction. PLOS ONE. Dataset. https://doi.org/10.1371/journal.pone.0039704",
"is_embargoed": false,
"embargo_date": null,
"embargo_type": null,
"embargo_title": "",
"embargo_reason": "",
"embargo_options": [],
"id": 123456,
"title": "Transcriptome Sequencing and Comparative Analysis of <em>Saccharina japonica</em> (Laminariales, Phaeophyceae) under Blue Light Induction",
"doi": "10.1371/journal.pone.0039704",
"handle": "",
"url": "https://api.figshare.com/v2/articles/123456",
"published_date": "2012-06-27T00:57:36Z",
"thumb": "https://s3-eu-west-1.amazonaws.com/ppreviews-plos-725668748/3074543/thumb.png",
"defined_type": 3,
"defined_type_name": "dataset",
"group_id": 107,
"url_private_api": "https://api.figshare.com/v2/account/articles/123456",
"url_public_api": "https://api.figshare.com/v2/articles/123456",
"url_private_html": "https://figshare.com/account/articles/123456",
"url_public_html": "https://plos.figshare.com/articles/dataset/Transcriptome_Sequencing_and_Comparative_Analysis_of_Saccharina_japonica_Laminariales_Phaeophyceae_under_Blue_Light_Induction/123456",
"timeline": {
"posted": "2012-06-27T00:57:36",
"firstOnline": "2016-01-19T09:09:40"
},
"resource_title": "Transcriptome Sequencing and Comparative Analysis of <em>Saccharina japonica</em> (Laminariales, Phaeophyceae) under Blue Light Induction",
"resource_doi": "10.1371/journal.pone.0039704"
}
Authenticate
Authentication is required for any endpoint that retrieves or accepts private or institutional information. A token can be created for any user account and provides access in line with the account’s privileges. In the example below, a user retrieves 10 basic metadata records from their personal account. These records may include both public and private (draft) records. Note that the results are limited to 10 by using the page and page_size parameters.
Python:
import json
import requests
#Set the base URL
BASE_URL = 'https://api.figshare.com/v2'
#Set the token for the header
api_call_headers = {'Authorization': 'token ENTER-TOKEN'} #example: {'Authorization': 'token dkd8rskjdkfiwi49hgkw...'}
#Retrieve basic metadata for 10 items your account owns
r=requests.get(BASE_URL + '/account/articles?page=1&page_size=10', headers=api_call_headers)
#Load the metadata as JSON
metadata=json.loads(r.text)
#View the metadata
print(metadata)
Output:
[
{
"id": 8365555,
"title": "Testing-full embargo-restrict access-unpublish-republish",
"doi": "10.0166/FK2.stagefigshareare.8365555",
"handle": "",
"url": "https://api.figshare.com/v2/account/articles/8365555",
"published_date": "2023-04-06T13:13:41Z",
"thumb": "",
"defined_type": 3,
"defined_type_name": "dataset",
"group_id": 9991,
"url_private_api": "https://api.figshare.com/v2/account/articles/8365555",
"url_public_api": "https://api.figshare.com/v2/articles/8365555",
"url_private_html": "https://figshare.com/account/articles/8365555",
"url_public_html": "https://faber.figshare.com/articles/dataset/Testing-full_embargo-restrict_access-unpublish-republish/8365555",
"timeline": {
"posted": "2023-04-06T13:13:41",
"firstOnline": "2023-04-06T13:09:40"
},
"resource_title": null,
"resource_doi": null
},
{
"id": 8356369,
"title": "My example dataset",
"doi": "10.0166/FK2.stagefigshareare.8356369",
"handle": "",
"url": "https://api.figshare.com/v2/account/articles/8356369",
"published_date": "2023-04-04T21:36:57Z",
"thumb": "",
"defined_type": 3,
"defined_type_name": "dataset",
"group_id": 9991,
"url_private_api": "https://api.figshare.com/v2/account/articles/8356369",
"url_public_api": "https://api.figshare.com/v2/articles/8356369",
"url_private_html": "https://figshare.com/account/articles/8356369",
"url_public_html": "https://faber.figshare.com/articles/dataset/My_example_embargoed_record_-_institutional_repository_version/8356369",
"timeline": {
"posted": "2023-04-04T21:36:57",
"firstOnline": "2023-04-04T21:36:01"
},
"resource_title": null,
"resource_doi": null
},
{
"id": 8351662,
"title": "Untitled Item",
"doi": "",
"handle": "",
"url": "https://api.figshare.com/v2/account/articles/8351662",
"published_date": null,
"thumb": "",
"defined_type": 0,
"defined_type_name": "",
"group_id": 9991,
"url_private_api": "https://api.figshare.com/v2/account/articles/8351662",
"url_public_api": "https://api.figshare.com/v2/articles/8351662",
"url_private_html": "https://figshare.com/account/articles/8351662",
"url_public_html": "https://faber.figshare.com/articles/dataset/_/8351662",
"timeline": {},
"resource_title": null,
"resource_doi": null
},
{
"id": 8332638,
"title": "Ant behaviour data set",
"doi": "10.0166/FK2.stagefigshareare.8332638",
"handle": "",
"url": "https://api.figshare.com/v2/account/articles/8332638",
"published_date": "2023-03-24T15:16:19Z",
"thumb": "https://s3-eu-west-1.amazonaws.com/testfigsharearepreviews.figshareare.com/830360616/thumb.png",
"defined_type": 3,
"defined_type_name": "dataset",
"group_id": 9991,
"url_private_api": "https://api.figshare.com/v2/account/articles/8332638",
"url_public_api": "https://api.figshare.com/v2/articles/8332638",
"url_private_html": "https://figshare.com/account/articles/8332638",
"url_public_html": "https://faber.figshare.com/articles/dataset/Ant_behaviour_data_set/8332638",
"timeline": {
"posted": "2023-03-24T15:16:19",
"firstOnline": "2023-03-24T15:16:19"
},
"resource_title": null,
"resource_doi": null
},
{
"id": 8329418,
"title": "A versioned item - unpublished - republished",
"doi": "10.0166/FK2.stagefigshareare.8329418",
"handle": "",
"url": "https://api.figshare.com/v2/account/articles/8329418",
"published_date": "2023-04-06T20:35:25Z",
"thumb": "https://s3-eu-west-1.amazonaws.com/testfigsharearepreviews.figshareare.com/830356944/thumb.png",
"defined_type": 2,
"defined_type_name": "media",
"group_id": 9991,
"url_private_api": "https://api.figshare.com/v2/account/articles/8329418",
"url_public_api": "https://api.figshare.com/v2/articles/8329418",
"url_private_html": "https://figshare.com/account/articles/8329418",
"url_public_html": "https://faber.figshare.com/articles/media/A_versioned_item_-_unpublished_-_republished/8329418",
"timeline": {
"posted": "2023-04-06T20:35:25",
"firstOnline": "2023-03-23T18:42:20"
},
"resource_title": null,
"resource_doi": null
},
{
"id": 8327074,
"title": "Data supporting wood ants behaviour",
"doi": "10.0166/FK2.stagefigshareare.8327074",
"handle": "",
"url": "https://api.figshare.com/v2/account/articles/8327074",
"published_date": "2023-03-22T09:57:21Z",
"thumb": "https://s3-eu-west-1.amazonaws.com/testfigsharearepreviews.figshareare.com/830355168/thumb.png",
"defined_type": 3,
"defined_type_name": "dataset",
"group_id": 9991,
"url_private_api": "https://api.figshare.com/v2/account/articles/8327074",
"url_public_api": "https://api.figshare.com/v2/articles/8327074",
"url_private_html": "https://figshare.com/account/articles/8327074",
"url_public_html": "https://faber.figshare.com/articles/dataset/Data_supporting_wood_ants_behaviour/8327074",
"timeline": {
"posted": "2023-03-22T09:57:21",
"firstOnline": "2023-03-22T09:57:21"
},
"resource_title": "A motion compensation treadmill for untethered wood ants (Formica rufa): evidence for transfer of orientation memories from free-walking training",
"resource_doi": "10.1242/jeb.228601"
},
{
"id": 8275510,
"title": "DRI Sample Report.pdf",
"doi": "10.0166/FK2.stagefigshareare.8275510",
"handle": "",
"url": "https://api.figshare.com/v2/account/articles/8275510",
"published_date": "2023-04-07T16:12:14Z",
"thumb": "",
"defined_type": 3,
"defined_type_name": "dataset",
"group_id": 9991,
"url_private_api": "https://api.figshare.com/v2/account/articles/8275510",
"url_public_api": "https://api.figshare.com/v2/articles/8275510",
"url_private_html": "https://figshare.com/account/articles/8275510",
"url_public_html": "https://faber.figshare.com/articles/dataset/DRI_Sample_Report_pdf/8275510",
"timeline": {
"posted": "2023-04-07T16:12:14",
"firstOnline": "2023-04-07T16:12:14"
},
"resource_title": null,
"resource_doi": null
},
{
"id": 8271566,
"title": "Exhibition B Image",
"doi": "10.0166/FK2.stagefigshareare.8271566",
"handle": "",
"url": "https://api.figshare.com/v2/account/articles/8271566",
"published_date": "2023-02-20T15:09:10Z",
"thumb": "https://s3-eu-west-1.amazonaws.com/testfigsharearepreviews.figshareare.com/830323572/thumb.png",
"defined_type": 2,
"defined_type_name": "media",
"group_id": 9991,
"url_private_api": "https://api.figshare.com/v2/account/articles/8271566",
"url_public_api": "https://api.figshare.com/v2/articles/8271566",
"url_private_html": "https://figshare.com/account/articles/8271566",
"url_public_html": "https://faber.figshare.com/articles/media/Exhibition_B_Image/8271566",
"timeline": {
"posted": "2023-02-20T15:09:10",
"firstOnline": "2023-02-20T15:09:10"
},
"resource_title": null,
"resource_doi": null
},
{
"id": 8262674,
"title": "Ireland pub C18th",
"doi": "10.0166/FK2.stagefigshareare.8262674",
"handle": "",
"url": "https://api.figshare.com/v2/account/articles/8262674",
"published_date": "2023-02-16T11:29:15Z",
"thumb": "https://s3-eu-west-1.amazonaws.com/testfigsharearepreviews.figshareare.com/830318778/thumb.png",
"defined_type": 1,
"defined_type_name": "figure",
"group_id": 9991,
"url_private_api": "https://api.figshare.com/v2/account/articles/8262674",
"url_public_api": "https://api.figshare.com/v2/articles/8262674",
"url_private_html": "https://figshare.com/account/articles/8262674",
"url_public_html": "https://faber.figshare.com/articles/figure/Ireland_pub_C18th/8262674",
"timeline": {
"posted": "2023-02-16T11:29:15",
"firstOnline": "2023-02-16T11:29:15"
},
"resource_title": null,
"resource_doi": null
},
{
"id": 8260098,
"title": "Slides",
"doi": "10.0166/FK2.stagefigshareare.8260098",
"handle": "",
"url": "https://api.figshare.com/v2/account/articles/8260098",
"published_date": null,
"thumb": "https://ndownloader.figshare.com/files/830317022/preview/830317022/thumb.png",
"defined_type": 7,
"defined_type_name": "presentation",
"group_id": 9999,
"url_private_api": "https://api.figshare.com/v2/account/articles/8260098",
"url_public_api": "https://api.figshare.com/v2/articles/8260098",
"url_private_html": "https://figshare.com/account/articles/8260098",
"url_public_html": "https://faber.figshare.com/articles/presentation/Slides/8260098",
"timeline": {},
"resource_title": null,
"resource_doi": null
}
]
Send data
Sending information through a POST or PUT endpoint is accomplished by adding a ‘data’ variable to the request. The contents of the data variable needs to be formatted as indicated by the documentation for the API endpoint. In the example below, a new record is added to the account that created the token.
Python:
import json
import requests
#Set the base URL
BASE_URL = 'https://api.figshare.com/v2'
#Set the token in the header
api_call_headers = {'Authorization': 'token ENTER-TOKEN'} #example: {'Authorization': 'token dkd8rskjdkfiwi49hgkw...'}
#Create json formatted for upload
sample_metadata = {"title":"Test metadata for upload","keywords":["biodiversity","invertebrate"]}
json_metadata = json.dumps(sample_metadata)
#Create a private item
r = requests.post(BASE_URL + '/account/articles', headers=api_call_headers, data = json_metadata)
if r.status_code == 201: #If Post was successful
print('Successfully created item')
Retrieve information for multiple items
This section describes how to use multiple API calls to retrieve metadata or files from multiple items. The basic idea is to:
- Create a list of item ids
- Loop through the list and gather the information needed for each item id
Retrieve item ids in your account
import json
import requests
#Retrieve list of private metadata for 50 items. This is for unpublished and published records.
#Set the base URL
BASE_URL = 'https://api.figshare.com/v2' <-------- Change figshare to figsh if working with the sandbox
#Set the token in the header
api_call_headers = {'Authorization': 'token ENTER-TOKEN'} #example: {'Authorization': 'token dkd8rskjdkfiwi49hgkw...'}
#Get items owned by account
r=requests.get(BASE_URL + '/account/articles?page=1&page_size=50', headers=api_call_headers)
items=json.loads(r.text)
if r.status_code != 200:
print('Something is wrong:',r.content)
else:
print('Collected',len(items),'metadata records')
#Create a list of all the item ids
item_ids = [item['id'] for item in items]
Retrieve item ids through a search query
This example searches for records with that use the category ‘Digital Humanities’ or ‘Environmental Humanities’
import json
import requests
BASE_URL = 'https://api.figshare.com/v2'
categories = ["'History and Philosophy of the Humanities'","'Environmental Humanities'"]
#Gather basic metadata for items (articles) that meet your search criteria
results = [] #create a blank list
for i in categories:
query = '{"search_for":":category: ' + i + '"}'
y = json.loads(query) #Figshare API requires json paramaters
#The number of results is unknown but you can collect up to 9,000 results. This sets the page size to 1000 results and calls the API
#to retrieve 9 pages of results
for j in range(1,10):
records = json.loads(requests.post(BASE_URL + '/articles/search?page_size=1000&page={}'.format(j), params=y).content)
results.extend(records) #add the retrieved records to the list of records
#See the number of items
print(len(results),'items retrieved')
#Create a list of all the item ids
item_ids_full = [item['id'] for item in results]
#Remove duplicates by converting to a dictionary and back to a list
item_ids = list( dict.fromkeys(item_ids_full) )
print(len(item_ids_full)-len(item_ids),'duplicate records removed,',len(item_ids),'unique records remain')
Retrieve item ids in a group
If you are at an institution (or not!), you may want to gather all the items in a particular group. The script below does just that. Note: It only returns items published in the specific group. It does not return results for subgroups
BASE_URL = "https://api.figshare.com/v2"
INST_ID = 319 #Institution ID, this example is for Iowa State University. You don't have to include this- Group is is enough
GRP_ID = 11962 #Group ID, this example is for the Iowa State University Agriculture Group.
#Gather basic metadata for items (articles) that meet your search criteria
results = [] #create a blank list
query = '{"institution": ' + str(INST_ID) + ',"group": ' + str(GRP_ID) + '}'
y = json.loads(query) #Figshare API requires json paramaters
#You can collect up to 9,000 results. This sets the page size to 1000 results and calls the API
#to retrieve 9 pages of results. You can use this endpoint to get the number of items in
#a group: https://docs.figshare.com/#stats_count_articles
for j in range(1,10):
records = json.loads(requests.post(BASE_URL + '/articles/search?page_size=1000&page={}'.format(j), params=y).content)
results.extend(records) #add the retrieved records to the list of records
#See the number of items
print(len(results),'items retrieved')
#Create a list of all the item ids
item_ids = [item['id'] for item in results]
Now gather information for those item ids
Full metadata
Use one of the methods above to create a list of item ids called item_ids. This script includes an option to add a token in the header in case some of the item ids are for unpublished records.
import json
import requests
#Set the token in the header if you have some private items in your list
#api_call_headers = {'Authorization': 'token ENTER-TOKEN'} #example: {'Authorization': 'token dkd8rskjdkfiwi49hgkw...'}
#---INSERT CODE TO COLLECT ITEM IDS HERE----
#Set the base URL
BASE_URL = 'https://api.figshare.com/v2'
full_records = [] #Create a blank list to hold the JSON metadata records
for id_value in item_ids:
r=requests.get(BASE_URL + '/account/articles/' + str(id_value), headers=api_call_headers)
metadata=json.loads(r.text)
full_records.append(metadata) #Add the collected metadata record to the master list of records
print(len(full_records),metadata records collected)
Views and Downloads
The options for views, downloads, and shares are described here: https://docs.figshare.com/#stats. Note that the endpoints for Breakdown and Timeline require a special administrator authentication that you can request for your institution through https://support.figshare.com.
In the example below, the total views and downloads for each item id are collected. Note that this script uses the pandas package to create a table of values.
import json
import requests
import pandas as pd
#---INSERT CODE TO COLLECT ITEM IDS HERE----
#Set the base URL
BASE_URL = 'https://stats.figshare.com'
stats = [] #This list will hold a dictionary for each item_id
for i in item_ids:
#make two api calls to retrieve total views and total downloads
r=requests.get(BASE_URL + '/total/views/article/'+ str(i))
views=json.loads(r.text)
r=requests.get(BASE_URL + '/total/downloads/article/'+ str(i))
downloads=json.loads(r.text)
#Add the item_id, views, and downloads as a dictionary and add to the stats list.
item_stats = {"item_id":i,"total_views":views['totals'],"total_downloads":downloads['totals']}
stats.append(item_stats)
#Convert the list of dictionaries to a dataframe
df = pd.DataFrame(stats)
#Show the top three rows of the dataframe
df.head(3)
Output:
| item_id | total_views | total_downloads |
|---|---|---|
| 13259588 | 243 | 54 |
| 1138718 | 7552 | 3030 |
Download file(s)
There are several ways to download files through the API. Each file that is part of a record has it’s own download URL. You can find this URL in the full metadata retrieved through the API or you can enter the item id at this endpoint: https://docs.figshare.com/#article_files. For item https://doi.org/10.6084/m9.figshare.5616409.v3, the download URL for the file is https://figshare.com/ndownloader/files/9778696. Visiting that URL will automatically start the download (File is 219KB).
Python:
import json
import requests
from pathlib import Path
#---INSERT CODE TO COLLECT ITEM IDS HERE----
#Set the base URL
BASE_URL = 'https://api.figshare.com/v2'
file_info = [] #a blank list to hold all the file metadata
for i in item_ids:
r = requests.get(BASE_URL + '/articles/' + str(i) + '/files')
file_metadata = json.loads(r.text)
for j in file_metadata: #add the item id to each file record- this is used later to name a folder to save the file to
j['item_id'] = i
file_info.append(j) #Add the file metadata to the list
#Download each file to a subfolder named for the article id and save with the file name
for k in file_info:
response = requests.get(BASE_URL + '/file/download/' + str(k['id']), headers=api_call_headers)
Path(str(k['item_id'])).mkdir(exist_ok=True)
open(str(k['item_id']) + '/' + k['name'], 'wb').write(response.content)
print('All done.')
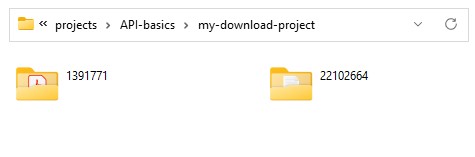
Output: The download script will save files to folders named for the item id the files belong to
Example workflows
Create an author report
The API can provide information for individual authors. As an example, we created a Jupyter Notebook in Google Colab.
Note The script retrieves statistics and if you want to do this for an account within an intsitutional repository, you need to adjust the stats urls in the script and will need your institution’s stats credentials. See the notes in the script. Also see the stats info in the ‘Advice and Guidance’ section at the top of the page.
Anyone can run this script, though you’ll need to sign into a Google account to do so. Then you must only add an author name or an ORCID. The script produces a table with metadata for the author’s outputs, views and downloads, and produces a plotly map of views for all the records (stats will only be for records stored in figshare.com accounts).
Here is the order of operations:
- Import libraries and set base API URL
- Perform search for name or ORCID using this endpoint: https://docs.figshare.com/#articles_search Then create a dataframe
- Gather views and downloads for each record from this endpoint: https://docs.figshare.com/#stats_totals Then create a dataframe
- Merge the metadata and stats dataframes
- Optionally collect all the same information for Collections using the collection endpoints
- Format the date information (extract dates from the JSON and add them to the dataframe
- Gather views by country for each item using this endpoint: https://docs.figshare.com/#stats_breakdown
- Map the views
- Save the dataframe and you can copy the map images from the browser
Create records, upload files, and publish
A common use of the API is to create metadata records from existing metadata. For example you may have metadata harvested from somewhere else that you want to include in your repository as metadata only or linked records. Or you may have metadata and files that you want to add to an account or repository. This section will assume you have already formatted the metadata for upload.
Create a metadata only or linked file record
To create records using the API, you need to use three endpoints: 1) create a private record, optionally add a link to a file, and then publish the record.
This Python example will create a linked file record for a paper- effectively it is a metadata only record that points users to the publisher’s DOI.
import json
import requests
#Set the token in the header and base URL
#Open a token stored in a text document:
text_file = open("symp-figsh-token.txt", "r") #Change this file name
TOKEN = text_file.read()
TOKEN.strip() #removes any hidden spaces
text_file.close()
#Or add your token as plain text (if you do, don't share this script with anyone):
#api_call_headers = {'Authorization': 'token ' + TOKEN}
#Set the base URL
BASE_URL = 'https://api.figshare.com/v2' #To use stage instance replace with 'https://api.figsh.com/v2'
#Create a json file with 1 record
jsonfile = {
"title": "Producing performance-advantaged bioplastics",
"description": "Abstract: A grand challenge for bio-based plastics is the ability to cost-effectively manufacture high-performance polymers directly from renewable resources that are also recyclable-by-design. A one-step conversion of xylose to polyesters has been reported, combining a sustainable lifecycle with impressive materials performance.",
"tags": [
"bioplastics"
],
"categories": [
614,
1058
],
"authors": [
{
"name":"Robin M. Cywar"
},
{
"id": "Gregg Beckham"
}
],
"defined_type": "journal contribution",
"license": 1,
"doi": "",
"handle": "",
"resource_doi": "10.1038/s41557-022-01030-y",
"resource_title": "Producing performance-advantaged bioplastics",
"timeline": {
"firstOnline": "2022-11-26"
},
"funding": "National Renewable Energy Laboratory",
"custom_fields": {
"Journal / Parent title": "Nature Chemistry",
"Volume": "14",
"Pagination":"967-969"
}
}
#You can also open a json file if you prefer:
#with open("beckham-record.json", "r", encoding='utf8') as read_file: #Replace this with the filename of your choice
# jsonfile = json.load(read_file)
#Upload the record
record_fails = []
jsonresult = json.dumps(jsonfile) #Takes one record and makes it a json string (double quotes)
r = requests.post(BASE_URL + '/account/articles', headers=api_call_headers, data = jsonresult)
if r.status_code != 201:
record_fails.append(str(r.content[0:75])) #Add failed index to list with partial description
else:
#Remove the admin account as an author by updating the record just created
#This uses the article url returned by the API response (r)
authordict = {}
authordict['authors'] = jsonfile['authors']
authorjson = json.dumps(authordict) #formats everything with double quotes
response_json = json.loads(r.content)
new_url = response_json['location']
s = requests.put(new_url, headers=api_call_headers, data = authorjson)
#Upload a link as a file
link = '{"link":"https://doi.org/'+ jsonfile['resource_doi'] +'"}'
tup = new_url.rpartition('/') #From end of URL, split string '/' so we can get the article id
article_id = str(tup[2]) #Then select the id from the resulting tuple.
t = requests.post(BASE_URL + '/account/articles/' + str(article_id) +'/files', headers=api_call_headers, data = link)
#Publish the record
u = requests.post(BASE_URL + '/account/articles/' + str(article_id) +'/publish', headers=api_call_headers)
#print(len(record_fails),"record fails. Failed record indexes:",record_fails)
print('The record is created and available here:',json.loads(u.content)['location'])
Upload files
The process to upload files may seem complex but this is necessary to handle extremely large files. So whether your file is a few kilobytes or a terabyte, this process will work.
The overall process to upload files looks like this: 1) create a private record, upload file(s), and then publish the record.
As described in the upload documentation, there are four steps to uploading: 1) Initiate the upload 2) Receive the number of file parts, 3) Upload the file parts, and 4) Complete the upload. The documentation linked above has a sample script in several languages. A Jupyter Notebook and Python example is downloadable from this Google Colab file and is reproduced below:
BASE_URL = 'https://api.figshare.com/v2/{endpoint}'
TOKEN = 'ENTER TOKEN BETWEEN QUOTES'
CHUNK_SIZE = 10485760 #about 10MB
FILE_PATH = 'FILE NAME' #If the file is in the same folder as this notebook, put the name of the file in quotes. E.g. 'file-name.mp4'
TITLE = 'Uploaded through API' #This is the title of the item you want to upload to.
#Define functions that split the file, initiate upload, and complete the upload
def raw_issue_request(method, url, data=None, binary=False):
headers = {'Authorization': 'token ' + TOKEN}
if data is not None and not binary:
data = json.dumps(data)
response = requests.request(method, url, headers=headers, data=data)
try:
response.raise_for_status()
try:
data = json.loads(response.content)
except ValueError:
data = response.content
except HTTPError as error:
print('Caught an HTTPError: {}'.format(error.message))
print('Body:\n', response.content)
raise
return data
def issue_request(method, endpoint, *args, **kwargs):
return raw_issue_request(method, BASE_URL.format(endpoint=endpoint), *args, **kwargs)
def list_articles():
result = issue_request('GET', 'account/articles')
print('Listing current articles:')
if result:
for item in result:
print(u' {url} - {title}'.format(**item))
else:
print(' No articles.')
def create_article(title):
data = {
'title': TITLE # You may add any other information about the article here as you wish.
}
result = issue_request('POST', 'account/articles', data=data)
print('Created article:', result['location'], '\n')
result = raw_issue_request('GET', result['location'])
return result['id']
def list_files_of_article(article_id):
result = issue_request('GET', 'account/articles/{}/files'.format(article_id))
print('Listing files for article {}:'.format(article_id))
if result:
for item in result:
print(' {id} - {name}'.format(**item))
else:
print(' No files.')
print
def get_file_check_data(file_name):
with open(file_name, 'rb') as fin:
md5 = hashlib.md5()
size = 0
data = fin.read(CHUNK_SIZE)
while data:
size += len(data)
md5.update(data)
data = fin.read(CHUNK_SIZE)
return md5.hexdigest(), size
def initiate_new_upload(article_id, file_name):
endpoint = 'account/articles/{}/files'
endpoint = endpoint.format(article_id)
md5, size = get_file_check_data(file_name)
data = {'name': os.path.basename(file_name),
'md5': md5,
'size': size}
result = issue_request('POST', endpoint, data=data)
print('Initiated file upload:', result['location'], '\n')
result = raw_issue_request('GET', result['location'])
return result
def complete_upload(article_id, file_id):
issue_request('POST', 'account/articles/{}/files/{}'.format(article_id, file_id))
def upload_parts(file_info):
url = '{upload_url}'.format(**file_info)
result = raw_issue_request('GET', url)
print('Uploading parts:')
with open(FILE_PATH, 'rb') as fin:
for part in result['parts']:
upload_part(file_info, fin, part)
def upload_part(file_info, stream, part):
udata = file_info.copy()
udata.update(part)
url = '{upload_url}/{partNo}'.format(**udata)
stream.seek(part['startOffset'])
data = stream.read(part['endOffset'] - part['startOffset'] + 1)
raw_issue_request('PUT', url, data=data, binary=True)
print(' Uploaded part {partNo} from {startOffset} to {endOffset}'.format(**part))
def main():
# We first create the article
list_articles()
article_id = create_article(TITLE)
list_articles() #Prints the list again and you should see your new item at the top of the list
list_files_of_article(article_id)
# Then we upload the file.
file_info = initiate_new_upload(article_id, FILE_PATH)
# Until here we used the figshare API; following lines use the figshare upload service API.
upload_parts(file_info)
# We return to the figshare API to complete the file upload process.
complete_upload(article_id, file_info['id'])
list_files_of_article(article_id)
#Run this to upload the file.
if __name__ == '__main__':
main()
Upload metadata and files in batch
If your existing metadata includes a URL or local location for the associated file(s) you can upload the metadata and files. Your existing metadata should be a list of dictionaries already formatted for upload (see the formatting page for advice). Use a for loop to upload the metadata for each record, then upload the file(s).
Batch format and upload metadata from a source
This offers an example of creating lnked file records based on harvested metadata. The resulting items clearly indicate the URL or DOI the user can click to find the original record. The example below is for paper metadata, but it can easily work with data metadata too. Macquarie University created a script to create catalog records in their repository from Dryad records.
Here are the steps to format metadata harvested from the Dimensions database and create linked file records:
- Open a json file (in the code below, an example file from Dimensions is manually created)
- Pull out the relevant fields and give them the proper keys (account for partial dates, author formatting, and missing abstracts)
- Convert the json record to a string with double quotes
- Upload the record
- Update the author list of the new record (removes the admin account as an author)
- Add the existing DOI as a linked file
This can upload to a specific group with specific custom metadata. You can change the api key to upload to different accounts or use impersonation.
Here is an example Python script. Note that this does not add Categories or Keywords which are required for publishing.
import json
import requests
import pandas as pd
#Set the token directly (don't share this notebook with this information)
TOKEN = "ENTER TOKEN HERE WITH QUOTES"
#Set the token in the header and base URL (change the file name and location)
#text_file = open("../../../testing-token-symlilliput.txt", "r")
#TOKEN = text_file.read()
#TOKEN.strip() #removes any hidden spaces
#text_file.close()
api_call_headers = {'Authorization': 'token ' + TOKEN}
#Set the base URL
BASE_URL = 'https://api.figshare.com/v2' #<--------- If using a sandbox, change this to 'https://api.figsh.com/v2'
#For demonstration, create a json file with 2 records- this is json formatted metadata from the Dimensions API.
#Feel free to change the information below so you can easily find these examples if using a shared sandbox
jsonfile = [
{
"abstract": "As robots are deployed to work in our environments, we must build appropriate expectations of their behavior so that we can trust them to perform their jobs autonomously as we attend to other tasks. Many types of explanations for robot behavior have been proposed, but they have not been fully analyzed for their impact on aligning expectations of robot paths for navigation. In this work, we evaluate several types of robot navigation explanations to understand their impact on the ability of humans to anticipate a robot’s paths. We performed an experiment in which we gave participants an explanation of a robot path and then measured (i) their ability to predict that path, (ii) their allocation of attention on the robot navigating the path versus their own dot-tracking task, and (iii) their subjective ratings of the robot’s predictability and trustworthiness. Our results show that explanations do significantly affect people’s ability to predict robot paths and that explanations that are concise and do not require readers to perform mental transformations are most effective at reducing attention to the robot.",
"authors": [
{
"affiliations": [
{
"city": "Pittsburgh",
"city_id": 5206379,
"country": "United States",
"country_code": "US",
"id": "grid.147455.6",
"name": "Carnegie Mellon University",
"raw_affiliation": "Carnegie Mellon University, Pittsburgh, PA, USA",
"state": "Pennsylvania",
"state_code": "US-PA"
}
],
"corresponding": "",
"current_organization_id": "",
"first_name": "Stephanie",
"last_name": "Rosenthal",
"orcid": [],
"raw_affiliation": [
"Carnegie Mellon University, Pittsburgh, PA, USA"
],
"researcher_id": "null"
},
{
"affiliations": [
{
"city": "Pittsburgh",
"city_id": 5206379,
"country": "United States",
"country_code": "US",
"id": "grid.147455.6",
"name": "Carnegie Mellon University",
"raw_affiliation": "Carnegie Mellon University, Pittsburgh, PA, USA",
"state": "Pennsylvania",
"state_code": "US-PA"
}
],
"corresponding": "",
"current_organization_id": "",
"first_name": "Peerat",
"last_name": "Vichivanives",
"orcid": [],
"raw_affiliation": [
"Carnegie Mellon University, Pittsburgh, PA, USA"
],
"researcher_id": "null"
},
{
"affiliations": [
{
"city": "Pittsburgh",
"city_id": 5206379,
"country": "United States",
"country_code": "US",
"id": "grid.147455.6",
"name": "Carnegie Mellon University",
"raw_affiliation": "Carnegie Mellon University, Pittsburgh, PA, USA",
"state": "Pennsylvania",
"state_code": "US-PA"
}
],
"corresponding": "",
"current_organization_id": "grid.147455.6",
"first_name": "Elizabeth",
"last_name": "Carter",
"orcid": [
"0000-0002-2735-148X"
],
"raw_affiliation": [
"Carnegie Mellon University, Pittsburgh, PA, USA"
],
"researcher_id": "ur.012471523754.61"
}
],
"date": "2022-12-31",
"doi": "10.1145/3526104",
"id": "pub.1147121755",
"title": "The Impact of Route Descriptions on Human Expectations for Robot Navigation",
"year": 2022
},
{
"abstract": "Data-driven network intrusion detection (NID) has a tendency towards minority attack classes compared to normal traffic. Many datasets are collected in simulated environments rather than real-world networks. These challenges undermine the performance of intrusion detection machine learning models by fitting machine learning models to unrepresentative “sandbox” datasets. This survey presents a taxonomy with eight main challenges and explores common datasets from 1999 to 2020. Trends are analyzed on the challenges in the past decade and future directions are proposed on expanding NID into cloud-based environments, devising scalable models for large network data, and creating labeled datasets collected in real-world networks.",
"authors": [
{
"affiliations": [
{
"city": "Pittsburgh",
"city_id": 5206379,
"country": "United States",
"country_code": "US",
"id": "grid.147455.6",
"name": "Carnegie Mellon University",
"raw_affiliation": "Carnegie Mellon University, Pittsburgh, PA",
"state": "Pennsylvania",
"state_code": "US-PA"
}
],
"corresponding": "",
"current_organization_id": "",
"first_name": "Dylan",
"last_name": "Chou",
"orcid": [],
"raw_affiliation": [
"Carnegie Mellon University, Pittsburgh, PA"
],
"researcher_id": "null"
},
{
"affiliations": [
{
"city": "Notre Dame",
"city_id": 8469294,
"country": "United States",
"country_code": "US",
"id": "grid.131063.6",
"name": "University of Notre Dame",
"raw_affiliation": "University of Notre Dame, Notre Dame, Indiana",
"state": "Indiana",
"state_code": "US-IN"
}
],
"corresponding": "",
"current_organization_id": "grid.131063.6",
"first_name": "Meng",
"last_name": "Jiang",
"orcid": [
"0000-0002-3009-519X"
],
"raw_affiliation": [
"University of Notre Dame, Notre Dame, Indiana"
],
"researcher_id": "ur.010456457135.66"
}
],
"date": "2022-12-31",
"doi": "10.1145/3472753",
"id": "pub.1141731091",
"title": "A Survey on Data-driven Network Intrusion Detection",
"year": 2022
}
]
#Format records for upload.
result = []
doi_list = []
for item in jsonfile:
my_dict={}
my_dict['title']=item.get('title')
if 'abstract' in item: #abstract isn't always present
my_dict['description']=item.get('abstract')
else:
my_dict['description']="No description available"
authors = [] #format authors
for name in item['authors']:
authorname = {"name" : name['first_name'] + " " + name['last_name']}
authors.append(authorname)
my_dict['authors']= authors
my_dict['defined_type'] = 'journal contribution'
my_dict['doi']= item.get('doi')
my_dict['resource_doi']= item.get('doi')
my_dict['resource_title']=item.get('title')
#my_dict['references'] = [item.get('URL')]
my_dict['timeline'] = {"firstOnline" : str(item['year']) + "-01-01"} #year only
#my_dict['is_metadata_record'] = True #Use these if you want a metadata only record
#my_dict['metadata_reason'] = 'See publisher version'
result.append(my_dict)
doi_list.append(item['doi'])
print(len(result),"records are ready for upload.")
#Upload the records
record_fails = []
partial_record_ids = []
created_record_ids = [] #Use this to delete all the draft records if needed
success_count = 0
count = 0 #This just tracks what index value the loop is on and is used to connect the metadata with the DOI update
for index, item in enumerate(result):
jsonresult = json.dumps(item) #Takes one record and makes it a json string (double quotes)
r = requests.post(BASE_URL + '/account/articles', headers=api_call_headers, data = jsonresult)
if r.status_code != 201:
record_fails.append(str(index) + ":" + str(r.content[0:75])) #Add failed index to list with partial description
count += 1
else:
count += 1 #increment here otherwise have to do it at each if statement below
#Remove the admin account as an author by updating the record just created
#This uses the article url returned by the API response (r)
# Get the location URL and item id
response_json = json.loads(r.content)
new_url = response_json['location']
item_id = response_json['entity_id']
created_record_ids.append(item_id)
#Format and update authors
authordict = {}
authordict['authors'] = item['authors']
authorjson = json.dumps(authordict) #formats everything with double quotes
s = requests.put(new_url, headers=api_call_headers, data = authorjson)
if r.status_code != 201:
record_fails.append(str(index) + "failed at author update:" + str(r.content[0:75])) #Add failed index to list with partial description
partial_record_ids.append(item_id)
else:
#Upload a link as a file
link = '{"link":"https://doi.org/'+ str(doi_list[count-1]) + '"}' #count-1 because already incremented value to next index
t = requests.post(new_url +'/files', headers=api_call_headers, data = link)
if r.status_code != 201:
record_fails.append(str(index) + "failed at doi update:" + str(r.content[0:75])) #Add failed index to list with partial description
partial_record_ids.append(item_id)
else:
success_count += 1
print(success_count,"records created and updated. There were",len(result)-len(created_record_ids),"records that were not created at all.")
print('There were',len(partial_record_ids),'records created but with a failed DOI link.')
print("Failed record descriptions:",record_fails)
Delete account items
You may have many draft items in your account, or if you are an admin, perhaps you’ve been testing with the Batch Management Tool, and you want to remove items.
Important Note: You cannot delete public items. For figshare.com users, you must contact Figshare to unpublish an item. For institution admins, this script provides a way to get a list of item ids to unpublish.
The following script uses a token from your account to get a list of both public and private/draft items in your account. If you only want to delete the current private items, you can pass the entire list of item ids to Figshare for deletion and Figshare will ignore requests to delete public items. If you are an admin who wants to delete everything, you’ll need to paste the list of item ids into your admin unpublish interface (in the Administration area). Figshare will unpublish the public items and then you can finish running the script to delete all the item ids.
import json
import requests
import pandas as pd
import csv
#Set the token directly (delete the lines above)
TOKEN = 'ENTER TOKEN BETWEEN SINGLE QUOTES'
api_call_headers = {'Authorization': 'token ' + TOKEN}
#Set the base URL
BASE_URL = 'https://api.figsh.com/v2' <---If using a production repository or figshare.com account change this to https://api.figshare.com/v2
#Now, gather records from your repository.
repository_items = []
num_pages = 1 #------> Change this as needed, but the code is set to gather 1000 records.
for i in range(1,num_pages+1):
item = json.loads(requests.get(BASE_URL + '/account/articles?page_size=1000&page={}'.format(i), headers=api_call_headers).content)
repository_items.extend(item)
print(len(repository_items),'metadata records collected')
df = pd.DataFrame(repository_items)
article_ids = df['id'].tolist()
print("If you need to unpublish items, paste these ids (no square brackets) in your admin interface:",article_ids)
#You may want to split this next block of code from the above blocks so that you can run it after unpublishing item ids.
delete_article_fails = []
for article in article_ids:
r = requests.delete(BASE_URL + '/account/articles/' + str(article), headers=api_call_headers)
if r.status_code != 204:
delete_article_fails.append(str(article) + ":" + str(r.content[0:75])) #Add failed index to list with partial description
print(len(delete_article_fails), 'errors!')
print('Fail descriptions:',delete_article_fails)
Retrieve metadata for items in review
There are several API endpoints that provide review/curation information. Reviewers can sort and inspect items in review through the GUI. But maybe you’d like to see a spreadsheet of the metadata for items that have a ‘pending’ status for review. This Google Colab notebook (also available as a Jupyter notebook) does just that. Be sure to use an admin token (not just a reviewer account token).
Here are the steps in that notebook:
- Retrieve a list of items with pending status in review
- Visit each item and retrieve all the metadata. Also add the owner name and email.
- Create a spreadsheet and do some formatting: dates, authors, add item owner, separate out custom fields.
- Save the spreadsheet
What if I want to publish those items?
You may want to bulk publish a group of items but avoid having to review them all. This isn’t possble through the API since that would defeat the purpose of review. However, you can use the Admin Batch Management tool to bulk publish items with review (making the assumption you reviewed the metadata in the CSV!). To do this you need to create a CSV with the item ids to publish. You can do this for pending review items that are in the spreadsheet from above. Here are the steps:
- Save a copy of the spreadsheet downloaded above as CSV (important: if using Excel, save as CSV rather than CSV UTF-8. The latter sometimes adds additional characters in the file)
- Delete all the items (rows) that you don’t want to publish
- Delete all columns except ‘id’
- Rename the ‘id’ column to ‘article_id’ and save the file
- You should now have a CSV with only one column of numbers titled ‘article_id’
- Log in to an administrator Figshare account that has review privileges.
- Navigate to the Batch Management tool and select your CSV as the source file. Select the option to automatically review items.
Impersonating user accounts
Impersonating accounts is described in this section of the documentation. Adding ‘impersonate=account_id’ to either the request URL for GET or in the body of a POST or PUT request will impersonate the account indicated. Here is a brief exaple that changes the author on an item owned by a user account and then publishes the item:
import json
import requests
#Set the token in the header and base URL
text_file = open("./././testing-token.txt", "r") #Paste your token in a text file and save it where this notebook is
TOKEN = text_file.read()
TOKEN.strip() #removes any hidden spaces
text_file.close()
api_call_headers = {'Authorization': 'token ' + TOKEN}
#Set the base URL
BASE_URL = 'https://api.figshare.com/v2' #Change this to 'https://api.figsh.com/v2' if you want to test in stage
#Get the author info from this endpoint: https://docs.figshare.com/#private_institution_accounts_list
new_user_id = 12345 #this is the user_id in the endpoint output
account_id = 6789 #This is 'id' in the endpoint output and is the account id for impersonation
#Set up the json to replace the author(s)
authors = {'authors': [{'id': new_user_id}], 'impersonate': account_id}
author_json = json.dumps(authors)
#Store the item id to be edited (you could also loop through a list of ids)
item_id = 65432
s = requests.put(BASE_URL + '/account/articles/' + str(item_id), headers=api_call_headers, data = author_json)
if s.status_code != 205:
print(str(s.content)) #print error
else:
#Publish the record
body = '{"impersonate":' + str(account_id) + '}' #impersonate to publish
u = requests.post(BASE_URL + '/account/articles/' + str(i) +'/publish', headers=api_call_headers, data = body)
if u.status_code != 201:
print(str(u.content)) print error
else:
print('Published!')
Create a repository dashboard
This is an example of how one might use the API to create a database for reporting purposes. The script downloads metadata from your repository along with views by item, views by country, and views by month. Eight tables are saved as worksheets within a Google Sheet. This sheet can be regularly updated so that any dashboards built off it will show current information.
Note: You need to have two sets of credentials:
- A repository administrator token
- The credentials for the stats API (ask Support if you do not have them yet)
The Jupyter Notebook is available and you can run it in Colab. Here are the high level steps in the process:
- Import libraries
- Set variables and metadata API credentials
- Retrieve all item ids and pull out only public item ids
- Retrieve full metadata and total views for each item id
- Create dataframes for each type of information: items, authors, funding, categories, and tags. Each dataframe includes the item id.
- Set variables and credentials for the stats API
- Use the stats API to retrieve views by country and over time.
- Either create a Google sheet or update an existing one
Visualize the data
You should be able to use the resulting spreadsheet as a data source for you favorite visualization software. We made a very basic proof of concept Looker Studio dashboard based on data from a live repository.
Search for records related to your institution
You can search for your institution’s name and do a high level analysis of the records out there. As an example, we created a Jupyter Notebook in Google Colab.
Anyone can run this script. One must only add search criteria and run all the cells in sequence. You can also save all the data as a Google sheet in your Google Drive.
Here is the order of operations:
- Import libraries and set base API endpoint and institution name
- Search using this endpoint: https://docs.figshare.com/#articles_search and create a dataframe
- Collect the full metadata for each item and create separate lists for item metadata, authors, funding, categories, tags, and files.
- Do some formatting and make dataframes from all the lists
- Start analysis by looking at the main dataframe columns
- Look at how many records are from institutions (using the existence of group_id)
- Item types with views
- Look at licenses
- See a list of linked funders (linked to Dimensions records)
- See grant titles
- See the top 20 tags
- See the top 20 categories
- See top 10 viewed items
- Export all the data to a Google sheet that you can then use for visualizations or other analysis
Custom Web Apps
You can create custom interfaces to display information from your Figshare account or repository. Online tools, like ChatGPT, can help. This was demonstrated in the API workshop Figshare ran at the 2023 Open Repositories Conference. You can see the resources for this demonstration on the workshop page and the workshop slides.